Abstract
Background: Peripheral T-cell lymphomas (PTCLs) are aggressive lymphomas with a wide range of clinical outcomes. Overall prognosis is worse for PTCL than B-cell lymphomas, but pts who achieve remission can be cured. After initial therapy, controversy exists regarding consolidation with autologous stem cell transplant, and clinical decisions are largely empirical. Circulating-tumor DNA (ctDNA) is emerging as a promising non-invasive biomarker that may aid clinical decision-making. Quantitative baseline levels of ctDNA and kinetics of ctDNA clearance during therapy may provide prognostic information. Further, novel methods that detect disease relapse prior to imaging may improve outcomes. We studied a high-throughput DNA sequencing method of detecting T-cell receptor (TCR) β and γ gene sequences in serum with a sensitivity of 1 lymphoma molecule per million leukocytes. Using this platform, we determined the predictive ability of monitoring ctDNA in pts undergoing frontline therapy for PTCL.
Methods: All pts had PTCL undergoing frontline EPOCH-based chemotherapy between May 1993 through December 2015. Pre-treatment FFPE biopsy samples and/or baseline serum samples were sent for VDJ amplification and TCR sequencing calibration. High-throughput TCR sequencing of the CDR3s of TCRβ/TCRγ genes was employed to provide a comprehensive and quantitative analysis of distinct T-cell clones within each sample, the relative frequency of each clone, and the exact unique nucleotide sequences of each clone's CDR3 regions. Pts with a calibrating TCRβ/TCRγ sequence had all available serum samples sent for identification and quantification. Prospectively collected serum sample time points included baseline, start of each cycle, and each follow-up visit until disease progression. Clinic visits after therapy included paired contrast CT scans performed at 3, 4, 6, 12 and 12 mos. intervals for the first 5 years, respectively. All analyses for ctDNA were performed blinded to clinical outcomes.
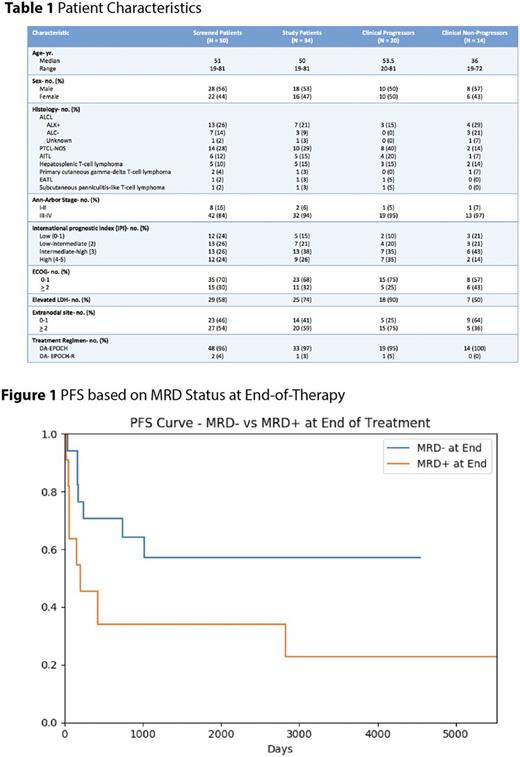
Results: Fifty PTCL pts had a pre-treatment FFPE or serum sample available for calibration. Nineteen of 23 (83%) pts with pre-treatment FFPE had a calibrating rearrangement; Fourteen of 27 (52%) pts with only pre-treatment serum available had a calibrating rearrangement; 1 pt who did not calibrate on FFPE had a dominant sequence identified in serum. Overall, tumor-specific clonotypes were identified in 34 (68%) of 50 pts tested. Ten of 18 (56%) pts with both FFPE and serum available had dominant clones in both compartments. Of the 44 calibrating pre-treatment samples, 1 calibrated for TCRβ only, 15 for TCRγ only, and 28 for both loci. Pt characteristics shown in Table 1. Of 34 study pts, 20 (59%) progressed and 14 (41%) were non-progressors. Quantitative baseline ctDNA levels were significantly higher in pts with stage IV disease compared to stages I-III (p=0.014) but did not correlate with LDH levels (p=0.53).
We analyzed the predictive ability of ctDNA as a measure of minimal residual disease (MRD) within 1 month of completing therapy in 28 pts. We compared pts who became MRD- to those who remained MRD+ and identified a trend towards improved PFS in those who became MRD- (median, NR vs. 208 days, p=0.07) (Figure 1). In 20 pts who achieved remission, we studied the use of ctDNA monitoring as a surveillance tool. Of the 10 pts who progressed after remission, ctDNA was detected prior to clinical relapse in 6 cases with a median lead-time of 13.2 mos. (range, 2.7 to 88.5) and 4 with detectable ctDNA > 12 mos. prior to clinical relapse. Four pts progressed without detectable ctDNA prior to clinical relapse. Interestingly, of the 10 non-progressors followed in surveillance, 5 pts developed low-level ctDNA positivity without clinical progression. Results of interim ctDNA kinetics and TCRβ testing is ongoing and will be presented at the meeting.
Conclusions: Next-generation based sequencing of the TCR can identify tumor-specific clonotypic sequences in the majority of pts from both FFPE and serum. As a marker of MRD, monitoring TCR sequences in serum may identify pts who do not benefit from treatment intensification, but further studies are needed. Surveillance monitoring of ctDNA in serum identifies TCR sequences months before clinical progression but not all pts with clonotypic sequences progressed clinically in our series. Further studies are needed to investigate surveillance monitoring methods and optimize clinical utility.
Jacob: Adaptive Biotechnologies: Employment, Equity Ownership. Chan: Adaptive Biotechnologies: Employment. Vignali: Adaptive Biotechnologies: Employment.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal